Links to code on Github: the singularizer and pluralizer, and the lemmatizer
Executive Summary:
I created a word lemmatizer in R and Python that automatically “digests” words down to their core form for the purpose of dimensionality reduction for NLP (Natural Language Processing) tasks. The lemmatizer works by recognizing formulaic endings to English words, yet handle enough special cases that they outperform stemmers such as Porter and Snowball in accuracy. Because this lemmatizer relies on structured word endings, it is less dependent on external dictionaries than most lemmatizers such as WordNet, and as such, it performs nearly equally as well on real or invented words that do not appear in dictionaries (unlike WordNet).
Motivations:
My motivations starting this project were simple: dig my hands deeper into NLP (Natural Language Processing) and develop useful code for myself where other code wouldn’t (or wouldn’t quickly) work. What started as a quick exploratory fix on my Wikipedia Spark project then turned into a substantial sub-project that carefully handles English words; I will focus on the final product here.
There are two main camps for dimensionality reduction in the NLP word: stemming and lemmatization. Stemmers (usually) simply chop off endings in an attempt to eliminate relatively meaningless differences in phonology (e.g. does the analysis really depend on whether he walks or I walk? The key idea is walk in both cases). While useful for their speed and simplicity, they suffer from an overly simplistic approach to manipulating words that results in many disconnected similar words (e.g., multiply and multiplication stem to similar but distinct terms, a type II error of sorts) as well as many spuriously connected words (e.g. universal, university, and the made-up word universion all stem to univers, a type I error of sorts). Lemmatizers, on the other hand, seek to capture the “lemma” or core/root of a word by looking through relationships in dictionaries. With lemmatizers, phonologically difficult relationships like that between better and good are accurately captured, so they’re very useful when accuracy is needed (especially for reduction of type I errors/overstemming). However, with a lemmatizer, one often has to put in extra under-the-hood work to link together related words of different parts of speech, first detecting the part of speech (with error) and then traversing the web of derivationally related forms (and synsets and pertainyms, etc.) until one decides on a “root” form. Furthermore, words not captured by the dictionary can’t be handled properly, which is a more common problem than one would expect. For instance, what’s the relationship between carmaker and carmaking? WordNet, one of the most powerful and popular lemmatizers (used by the popular NLTK package), doesn’t know, as carmaking isn’t defined. Undefined words can be accurately processed by stemmers (the carmaking example is, for example) but pose a real problem for many lemmatizers. Clearly there is a middle ground between the overly general world of stemming and overly specific world of lemmatizing, so I set out to create a program that would find it.
English as a language is characterized by its decentralization and archaic spelling as well as its repeated (though occasionally inconsistent) use of patterns to generate inflectional forms. This fact requires a mix of dynamic/rule-based approaches (to generalize the inflectional system) as well as static/case-based approaches (to successfully handle legacy spellings and manage the confusing mixture of Latin, Greek, French, and Anglo-Saxon morphology that characterize English words).
So, I created the “word digester” to combine the best aspects of stemmers (simplicity/ease of use; usefulness on words left out of the dictionary) with the best aspects of lemmatizers (comprehensiveness; handling of special cases; readability of outputs). It will take any English-like word (newly invented or well-established) and digest it into its core form. It handles enough special cases to be useful on some of the more peculiar English word forms, yet it uses enough general rules to be useful without relying on a dictionary. It’s not prohibitively slow either (digesting a 20k-word dictionary in roughly 1-3 seconds in both R and Python). Admittedly, as a recent personal project of definite complexity, there are still plenty of gaps in its coverage of special cases, which can be fixed in time. Below I’ll highlight a few examples that highlight the capabilities of the word digester and also discuss the singularizer and pluralizer functions I created as a companion project to the digester.
The (real or invented) words multiple, multiples, multiply, multiplied, multiplies, multiplier, multipliers, multiplying, multiplication, multiplications, multiplicative, multiplicatives, multiplicatively, multiplicativity, multiplicativeness, multiplicationment, multiplicativement, antimultiplicative, unmultiplicative, nonmultiplicative, dismultiplicative, multiplicate, multiplicating, and potentially more all reduce to multiple.
It handles some of the most relevant Latinate or Greek plurals as well: symposia and symposiums both reduce to symposium, and lemmata and lemmas both reduce to lemma, for instance, and the pluralize function allows selective creation of either plural form in the reverse direction. In addition, hypotheses reduces to hypothesis, matrices to matrix, series to series, spectra to spectrum, phenomena to phenomenon, alumni to alumnus, and so on.
Anglo-Saxon-style forms are also covered: berries reduces to berry, cookies to cookie, leaves to leaf, children to child, oxen to ox, elder to old, (wo)men to (wo)man, people to person, brethren to brother, and so on.
Many irregular changes are covered: pronunciation reduces to pronounce, exemplify to example, redemption to redeem, tabular to table, operable (and the made-up but more regular-looking operatable) to operate, despicable to despise, grieve to grief, conception to conceive, flammable to flame, statuesque to statue, wholly to whole, shrubbery to shrub, and so on.
Sensible words outside the dictionary are also properly handled: both buttery as well as butterific reduce to butter, and planning as well as the E.U.-favored planification both reduce to plan. Feel free to make up your own words and try them; currently only some rules are sufficiently generalized, so while antidisestablishmentarianismesquely reduces to establish, I can’t promise that every made-up word will be processed correctly.
I’ve verified, semi-manually, the digester’s results on a few corpora: a 1k-word corpus, a 20k-word corpus, and a 370k-word corpus. No errors were made with the 1k corpus; nearly all of the 1200 or so purported errors (defined here as digested word results that do not also appear in the same source corpus) made on the 20k corpus are either—from most common to least common—errors on proper nouns (expressly not intended to be handled by the function as other methods exist for that purpose), real words that do not appear in that corpus (not an error), or genuine errors that I haven’t bothered to fix yet (estimated on the order of 10 or so). This implies a true error rate of around 1% or less on the 20k-word corpus. I have skimmed through the purported errors on the 370k-word corpus (~47k of them), and have similarly found that most of the purported errors either are on proper nouns, are not real errors at all, or are on exceptionally rare, esoteric, often medicinal or taxonomical words (as these make up most of the 370k-word corpus to begin with as a consequence of the Zipf distribution of word frequency).
How does it work? Take a peek at the code on Github to see for yourself—though it is long, it is well-commented and fairly self-explanatory. The basic process involves handling each of the many recognized suffixes in the order in which they almost universally appear in words. Special cases are handled before the best-guess set of general rules handle the suffixes. Masks are used to hide words that have suffix-like endings that should not be removed. One of the most difficult aspects of making the digester as accurate as possible was designing an effective regex test to determine whether an ultimate “e” should remain attached to a word after suffix removal. The suffixes are handled in groups, in which only relevant subsets of words (those that end with something that looks like the suffix in question) are handled for efficiency. Writing some aspects of this code in C would certainly greatly speed up computation, but there isn’t any need for that on most scales as is, as I’ve designed the code such that it does not scale directly with the size or length of the input, but rather with the cardinality of the input (i.e. it handles each unique word only once).
The order I’ve used for processing is:
- singularize plural forms
- remove contractions
- fix irregular common words (all as special cases)
- remove certain prefixes (with a mask to prevent similar non-prefix forms from being removed)
- remove adverbial ly suffix
- remove noun-form ness and ity suffixes
- remove adjectival esque and ish suffixes
- remove adjectival able and ible suffixes
- remove noun-form hood suffix
- edit miscellaneous Saxon- and French-style suffixes
- remove istic, ist, and ism suffixes
- remove adjectival al, ial, ical, ual, and inal suffixes
- remove adjectival ian suffix
- remove adjectival ary suffix
- remove noun-form ment suffix
- remove adjectival ic suffix
- remove adjectival ous suffix
- remove adjectival ful and less suffixes
- remove adjectival ar and iar suffixes
- edit noun-form ance and ence suffixes
- remove adjectival ant and ent suffixes
- remove adjectival ive suffix
- remove noun-form age suffix
- remove noun-form tion and ion suffixes
- remove noun-form ery suffix
- remove adjectival y suffix
- remove noun-form itude suffix
- edit noun-form ysis suffix
- remove comparative and doer-form er suffix
- remove superlative est suffix
- remove verb-form ed suffix
- remove verb-form ing suffix
- remove verb-form ify suffix
- remove verb-form ize suffix
- remove verb-form ate suffix
- fix miscellaneous verb forms to similar noun forms (e.g, multiply to multiple)
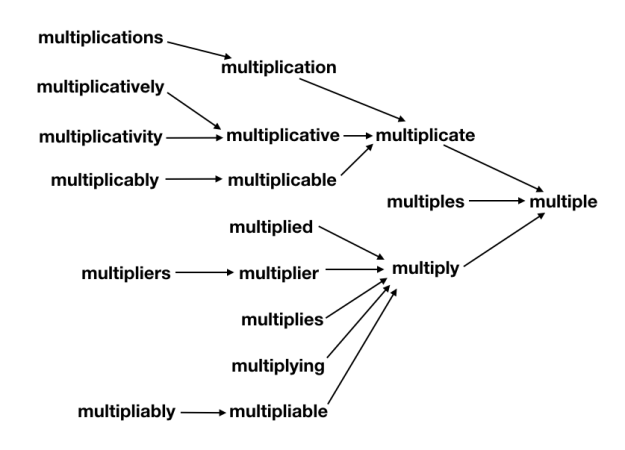
What’s important to note is that each step along the way, the suffixes are removed such that a valid word is produced. In this way, the transformation pipeline functions such that it “collects” different derivationally related forms on their way toward the base form, analogously to how a river’s tributary system works. As such, any failure of the system to properly join one subset of derivationally related forms into further pathways toward the base form will leave that subset as a single valid (albeit not completely basal) word instead of creating something new and unrecognizable. Additionally, this design makes debugging and adding extra special cases easy, as each special case only need be handled once in order for all its derivationally related forms to also be successfully processed. Similarly, it’s straight-forward to force the system to not join together some branches if desired––the change need only be made in one place.
Coda:
I initially coded up these functions in R, though wanting to expose it to wider use, especially within the NLP universe (which is very Python-centric), I transcribed the code into Python with the help of an R-to-Python transcription program I wrote myself. To learn more about that process, see this post.